An emission inventory is built and reported for a variety of pollutants, such as CO, NOx, VOC, PM10, and SO2. However, AQM chemical mechanisms (e.g., CB4) contain a simplified set of equations that use “model species” to represent atmospheric chemistry. Therefore, emissions processing requires speciation profiles to convert the emissions in terms of pollutant values to the species used in the photochemical mechanism. The purpose of the chemical speciation processing program Spcmat is to produce matrices that contain the factors for converting the input emissions pollutants to the model species used in the AQM. These species include organics, PM species, and toxics species.
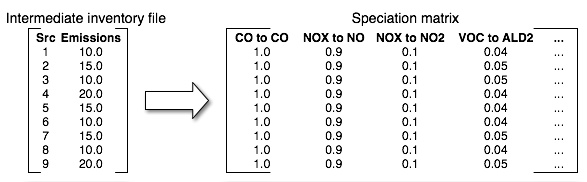
The speciation matrices that Spcmat creates are used transform column vectors of inventory-pollutant emissions into column vectors of model-species emissions. As shown in Figure 2.21, “Relationship of inventory sources to speciation matrix”, the speciation matrix consists of columns for each required pollutant-to-species transformation and includes an entry for each source. The entries are the factors needed to convert the inventory pollutants into the model species. Note that speciation matrices depend only upon the chemical mechanism and the inventory, and they are therefore independent of the other factor-based operations for emissions processing.
Chemical speciation processing addresses the following issues during emissions processing:
-
Splitting inventory pollutants into chemical species
-
Pollutant-to-pollutant conversions
-
Checking the consistency of the speciation profiles with the inventory
-
Setting the order of the output species
-
Creating speciation intermediate files
In the subsections below, we provide additional detail about each of these steps, in the order in which they are listed above.
SMOKE supports run-time, user-selected inventory pollutants and chemical mechanisms. Before running the chemical speciation step, the only relevant information that SMOKE has is information about the inventory pollutants. After Spcmat runs, SMOKE then has the instructions for supporting a specific chemical mechanism, and through the speciation matrices, SMOKE will be able to generate model-ready emissions for the specific chemical mechanism set by the user. The inventory pollutants relate to the chemical mechanism because certain pollutants are needed to create certain species, but the pollutants do not dictate the chemical mechanism. As a SMOKE user, you must be aware of what pollutants are required to generate the model species needed by a chemical mechanism, so that all needed model species are created.
As we just mentioned, SMOKE learns of the species being created for a given run through the Spcmat program. The chemical speciation profiles input file (GSPRO) is the data file that controls the chemical species SMOKE will create. It contains the chemical speciation profile code,
the pollutant-to-species relationships, and both mole-based and mass-based conversion factors. The format and contents of
this file are described in detail in Section 8.5.2, “GSPRO: Speciation profile file”. Several GSPRO files to support many chemical mechanisms are available for download; see Section 4.2, “Test case descriptions” for more information. How to select an existing chemical mechanism or specify a new one, and how to ensure that the GSPRO file is consistent with other input files, are described in Section 4.4.10, “Use a different speciation mechanism or change speciation inputs”.
Spcmat uses a cross-reference file to assign the chemical speciation profiles to the inventory sources and pollutants. The cross-reference
file can assign the profiles using a hierarchy that is based on the source characteristics; a detailed list of valid assignments
is given in Section 6.15, “Spcmat”. All assignments are pollutant-specific, such that each pollutant for a source can (and often should) use a different speciation
profile. The cross-reference file can also assign a default profile, and some pollutants that have only one way of being speciated
(e.g., mapping the CO pollutant to the CO species) receive a default profile for every source. When Spcmat makes default assignments, you can optionally choose to have warnings given for all of these assignments using the REPORT_DEFAULTS option. Spcmat will also produce a warning about any inventory pollutants that are not assigned a speciation profile, because this will
result in the emissions for that pollutant being dropped by SMOKE.
Spcmat creates two speciation matrices during each run: a mole-based matrix and a mass-based matrix. The speciation profile file
(GSPRO) has different factors for mass- and mole-based conversions. It is not trivial to convert between mass- and mole-based factors
for some chemical mechanisms like CB4, which use aggregates of chemical compounds or parts of compounds to define the model
species. One cannot simply use a molecular weight to convert accurately, because the molecular weight of the chemical species
is different for every speciation profile. This is because different proportions of chemical compounds are present in each
speciation profile, so even though the species are the same, their molecular weights are different from profile to profile.
This is why SMOKE has the two speciation matrices. The mole-based matrices are used to create the model-ready files, and the
mass-based matrices are used only to create the reports that require tons, kilograms, grams, or other mass units. One peculiarity
of the mole-based matrix is that particulate species emissions cannot be expressed in moles, so the units are still grams
in the mole-based matrices for particulate species.
Chemical speciation has both similarities and differences from the aggregation and disaggregation that is performed during inventory import (see Section 2.9.7, “Aggregate or disaggregate toxics emissions”). It is similar in that it involves separating one data value into more than one data value. For example, inventory disaggregation can split Mercury & Compounds (CAS=199) into elemental mercury, divalent gaseous mercury, and divalent particulate mercury during inventory import of nonroad mobile sources. Similarly, NOx can be split into nitrogen oxide (NO) and nitrogen dioxide (NO2) during chemical speciation. Aggregation is also similar to speciation because it can map multiple pollutants or parts of pollutants into the same chemical compound, just as speciation can map parts of the different pollutants (e.g., HGSUM and HG) to the same model species (e.g., HG). On the other hand, the two concepts are different for three reasons. First, the pollutants aggregated and disaggregated by Smkinven are still considered by SMOKE to be pollutants, not species, to pass to later processing steps. Second, chemical speciation allows multiple split factors to be used for the same pollutant-to-species conversion, whereas inventory aggregation/disaggregation uses just one factor across the whole inventory for each pollutant-to-pollutant conversion. Third, chemical speciation also converts the units of the pollutants (tons) to the units of the species (moles for gaseous species and grams for particulate species).
Speciation profiles do not necessarily conserve mass. For example, it is possible to input 100 tons of VOC into the Spcmat program and have it output factors that will produce 70 tons of VOC-based species or 110 tons of VOC-based species. The reduced mass occurs when some of the pollutant’s mass does not map to chemically reactive species in the inventory. In some cases, the nonreactive (NR) species is included in the speciation profiles so that the speciation profiles do sum to 1. Increased mass happens because some compounds that are part of VOC may have chemical reactivity associated with two model species. Since this one part of the VOC is mapped to two model species, its mass appears to be double-counted when summing the model-species mass. This is merely an artifact of how the model species are defined and implemented in the AQM, and the AQM is responsible for accounting for such issues in its chemical mechanism.