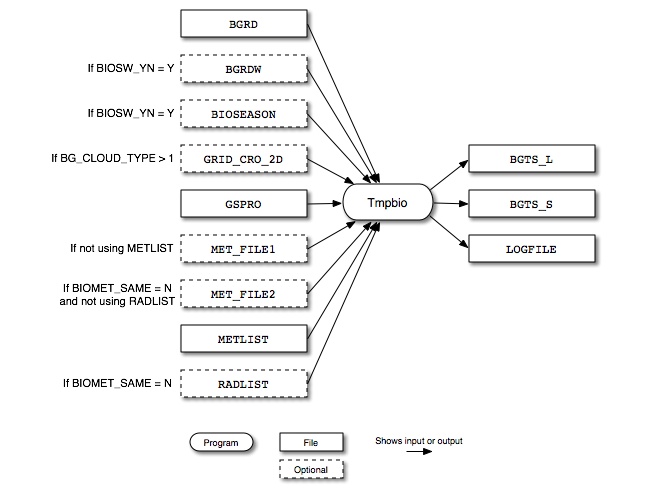
Figure 6.27, “Tmpbio input and output files” shows the input and output files for the Tmpbio program. The input files include the gridded, normalized biogenic emissions from Rawbio (BGRD and optionally BGRDW) and, if BIOSW_YN = Y, an optional gridded seasonal switch file output from Metscan (BIOSEASON). Rawbio also reads the optional meteorology file GRID_CRO_2D and the speciation profiles file (GSPRO). For inputting surface pressure and radiation/cloud data, Tmpbio can either use individual meteorology files (MET_FILE1 and MET_FILE2) or list files (METLIST and RADLIST).
Tmpbio outputs gridded, speciated, hourly biogenic emissions files in moles/hour (BGTS_L) and tons/hour (BGTS_S) and a log file detailing the program’s execution (LOGFILE).
| File Name | Format | Description |
|---|---|---|
BGRD |
I/O API NetCDF | Gridded, normalized summer biogenic emissions, output from Rawbio |
BGRDW |
I/O API NetCDF | Gridded, normalized winter biogenic emissions, output from Rawbio |
BIOSEASON (optional)
|
I/O API NetCDF | Gridded seasonal switch file containing daily data, with “0” meaning use winter normalized emissions and “1” meaning use summer normalized emissions, produced by Metscan |
GRID_CRO_2D (optional)
|
I/O API NetCDF | Meteorology file containing latitude and longitude coordinates for each grid cell |
GSPRO |
ASCII | Speciation profiles file |
MET_FILE1 |
I/O API NetCDF | Meteorology file containing temperature and surface pressure data and possibly radiation/cloud data as well (see BIOMET_SAME environment variable)
|
MET_FILE2 (optional)
|
I/O API NetCDF | Meteorology file containing radiation/cloud data if not in MET_FILE1 |
METLIST |
ASCII | List of meteorology files containing temperature and surface pressure data and possibly radiation/cloud data as well (see
BIOMET_SAME environment variable)
|
RADLIST (optional)
|
ASCII | List of meteorology files containing radiation/cloud data if not in the files listed by METLIST |
-
BG_CLOUD_TYPE: [default: 1]Specifies which meteorological data to use.
-
1: Use RGND or RSD (solar radiation reaching the ground) from MM5 to calculate PAR
-
2: Use MM5 Kuo cloud fractions and surface pressure data to calculate PAR (untested)
-
3: Use MM5 Kain-Fritsch cloud fractions and surface pressure data to calculate PAR (untested)
-
4: Use MM5 cloud fractions with no deep convection parameterization and surface pressure data to calculate PAR (untested)
-
5: Assume clear skies when calculating PAR; need
GRID_CRO_2Dfile (latitude-longitude coordinates) for solar zenith angle calculation
-
-
BIOG_SPRO: [default: 0000]Specifies the speciation profile for biogenic emissions from
GSPRO. -
BIOMET_SAME: [default: N]Indicates whether the radiation/cloud data are in the same file as the temperature data (
MET_FILE1) or a different file (MET_FILE2). -
BIOSW_YN: [default: TRUE]Indicates whether to input a gridded seasonal switch file.
-
OUTZONE: See description in Section 6.17.3.2, “Input Environment Variables” -
PRES_VAR: [default: PRSFC]Specifies the variable name for surface pressure data to extract from
MET_FILE1. -
RAD_VAR: [default: RGRND]Specifies the variable name for radiation/cloud data to extract from either
MET_FILE1or optionalMET_FILE2. -
TMPR_VAR: [default: TA]Specifies the variable name for temperature data to extract from
MET_FILE1.