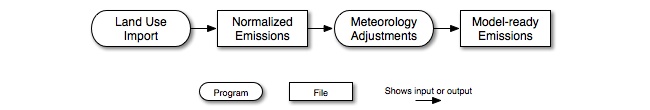
SMOKE biogenic emissions modeling can be accomplished with two different approaches to the science used. The first is the Biogenic Emissions Inventory System, version 2 (BEIS2) approach, which uses an adaptation of the BEIS2 model that fits within the data flows of SMOKE and works with the meteorology and AQMs used with Models-3. The second approach uses version 3.09 or version 3.14 of BEIS. For either approach, the processing scheme is the same (Figure 2.11, “Biogenic-source processing steps and intermediate files”). The raw land use inventory data are imported and output as normalized emissions. Meteorology adjustments are then applied to the normalized emissions to create hourly model-ready emissions estimates.
For BEIS2 processing, the land use import can start with either the county-total land use data or gridded land use data. In
both cases, the land use import step produces the same gridded normalized emissions output file (BGRD), which contains gridded time-independent emissions for groups of land use biomass data. The emissions are computed using
emission factors from either a summer or winter emission factor lookup table, which are specific to BEIS2. BEIS2 processing
uses the BELD2 land use categories. It is also possible to use the BELD3 land use data in BEIS2 processing by applying the
Beld3to2 SMOKE utility described in Section 5.3.3, “Beld3to2”. Meteorology adjustments and a single chemical speciation profile are applied to these normalized emissions to calculate
model-ready emissions.
For BEIS3 processing, the land use import can start only with gridded BELD3 land use data and uses BEIS3 summer and winter emission factors. In Section 2.17, “Biogenic processing”, we provide additional details about the SMOKE programs used for BEIS2 and BEIS3 processing and their capabilities.
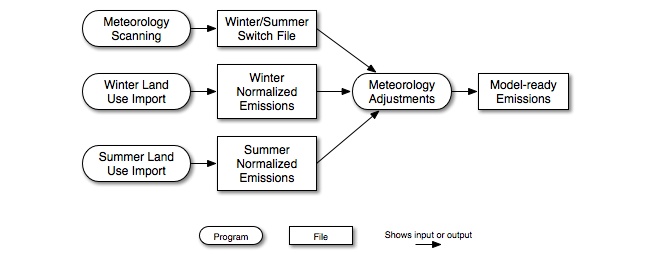
For both BEIS2 and BEIS3, a variation can be run on the processing steps shown in Figure 2.11, “Biogenic-source processing steps and intermediate files” (see Figure 2.12, “Biogenic-source processing steps and intermediate files using both winter and summer emission factors”). In this variation, some grid cells use summer emission factors and some use winter emission factors. This is useful during the changes of seasons. Based on guidance from EPA, the summer emissions factors should be used for time periods after the last frost of the spring until the first frost of the fall, and winter emission factors should be used at other times of the year. To make such assignments by grid cell, the SMOKE utility Metscan analyzes the meteorology data for the entire year (or the period of interest) to establish which days each grid cell should use winter and summer emission factors. Metscan creates a winter/summer switch file that indicates the appropriate season for each grid cell for each day. More information on Metscan is available in Section 5.3.11, “Metscan”. The results of the meteorology analysis can then be used in the Figure 2.12, “Biogenic-source processing steps and intermediate files using both winter and summer emission factors” processing approach, in which both the summer and winter normalized emissions are provided to the meteorology adjustments step, along with the winter/summer switch file. The resulting model-ready emissions data have used the winter emission factors for all grid cells of the domain that have experienced the first freeze date of the year but not the last (within a calendar year, this is the time periods January through March and November through December in many regions), and the summer emission factors for all grid cells between the last and first freeze dates.
Figure 2.12. Biogenic-source processing steps and intermediate files using both winter and summer emission factors
In Section 2.17, “Biogenic processing” we describe the SMOKE programs that are needed for each of these processing types for both BEIS2 and BEIS3 processing, and additional details about what activities are accomplished during each step.